Ligands of CASF-2016
Published:
CASF-2016 is a commonly used benchmark for docking tools. Unfortunately, some of the provided ligand files cannot be loaded using RDKit (version 2022.09.1) but there is an easy remedy.
The ligands are provided in two file formats – mol2 and sdf. Let us try reading the provided sdf files first.
# load CASF-2016 SDF files with RDKit
from pathlib import Path
from rdkit.Chem.rdmolfiles import SDMolSupplier
path_casf = Path('./CASF-2016/coreset')
names = sorted([d.stem for d in path_casf.iterdir() if d.is_dir()])
success = set()
failed = set()
for name in names:
path_sdf = path_casf / name / f"{name}_ligand.sdf"
mols = SDMolSupplier(str(path_sdf), sanitize=True)
if len(mols) > 0 and mols[0] is not None:
success.add(name)
else:
failed.add(name)
print("Success:", len(success))
print("Failed:", len(failed))
Running the above we get 86 failures for 285 files.
Let us try the provided mol2 files next.
# load CASF-2016 MOL2 files with RDKit
from rdkit.Chem.rdmolfiles import MolFromMol2File
success = set()
failed = set()
for name in names:
path_mol2 = path_casf / name / f"{name}_ligand.mol2"
mol = MolFromMol2File(str(path_mol2), sanitize=True)
if mol is not None:
success.add(name)
else:
failed.add(name)
print("Success:", len(success))
print("Failed:", len(failed))
print(sorted(failed))
This time we only get 12 failures.
If we use the mol2 files first and fall back to the SDF file, we get 6 ligands which we cannot read properly. They are the ligands for complexes 1BZC, 1VSO, 2ZCQ, 2ZCR, 4TMN, and 5TMN.
To see what is going on, we spot check the sdf file of 5TMN. The sanitization error reads “explicit valence for atom # 25 C, 6, is greater than permitted”.
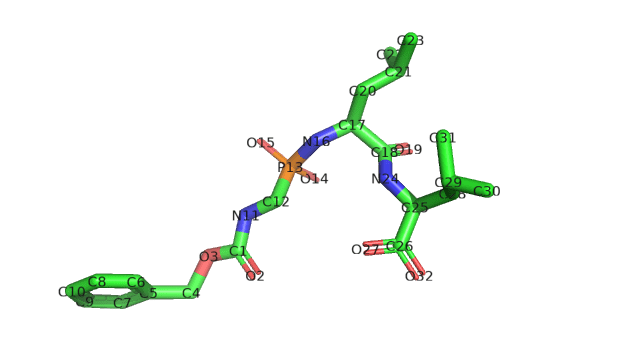
The mol2 files with error message “warning – O.co2 with non C.2 or S.o2 neighbor.”
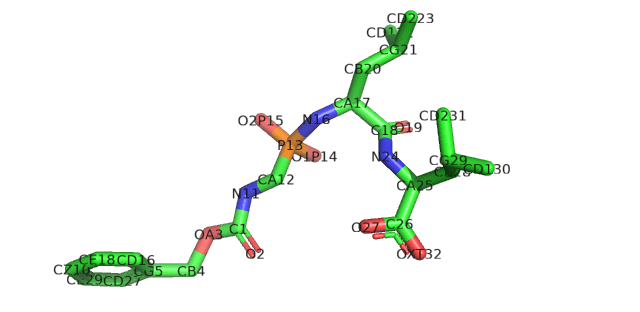
The easiest way to solve these errors is to go find the ligand in the PDB and download a new sdf file from there. Viola, this time the file can be read, and we get a nice ligand.
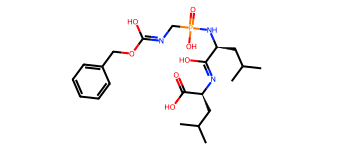
Luckily we only have to do download a new file 6 times.
