Bad chemistry in old protein-ligand binding complex data set
Published:
The Astex Diverse set1 is a dataset containing the crystallized poses of 85 protein-ligand complexes. It was introduced in 2007 to address problems in previous datasets such as incorrect ligand representation.
Loading the 85 ligand files with today’s version of the cheminformatics toolkit RDKit2 is, however, not as straightforward as you might expect.
29 out of the 85 files fail RDKit’s sanitization checks because they contain neutral nitrogen atoms connected to two carbons and two hydrogens. The problem is that the RDKit does not automatically add the missing positive charge when loading the files.
Going back to the original source of these structures, the PDB, we can obtain files that can be loaded by the RDKit without sanitization errors but they do not contain the same protonation pattern.
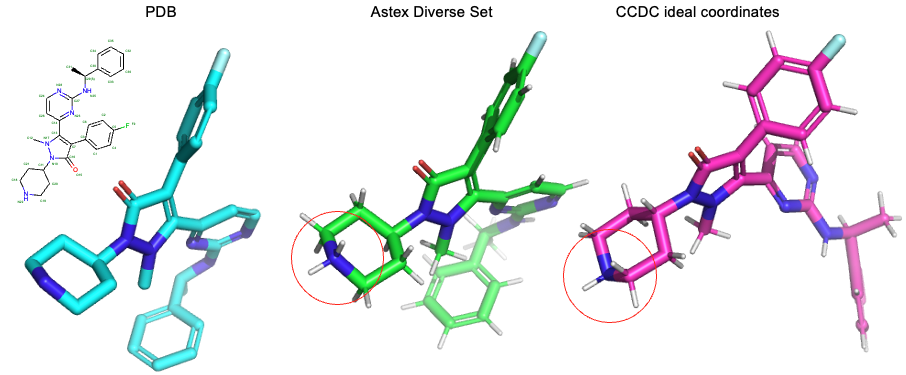
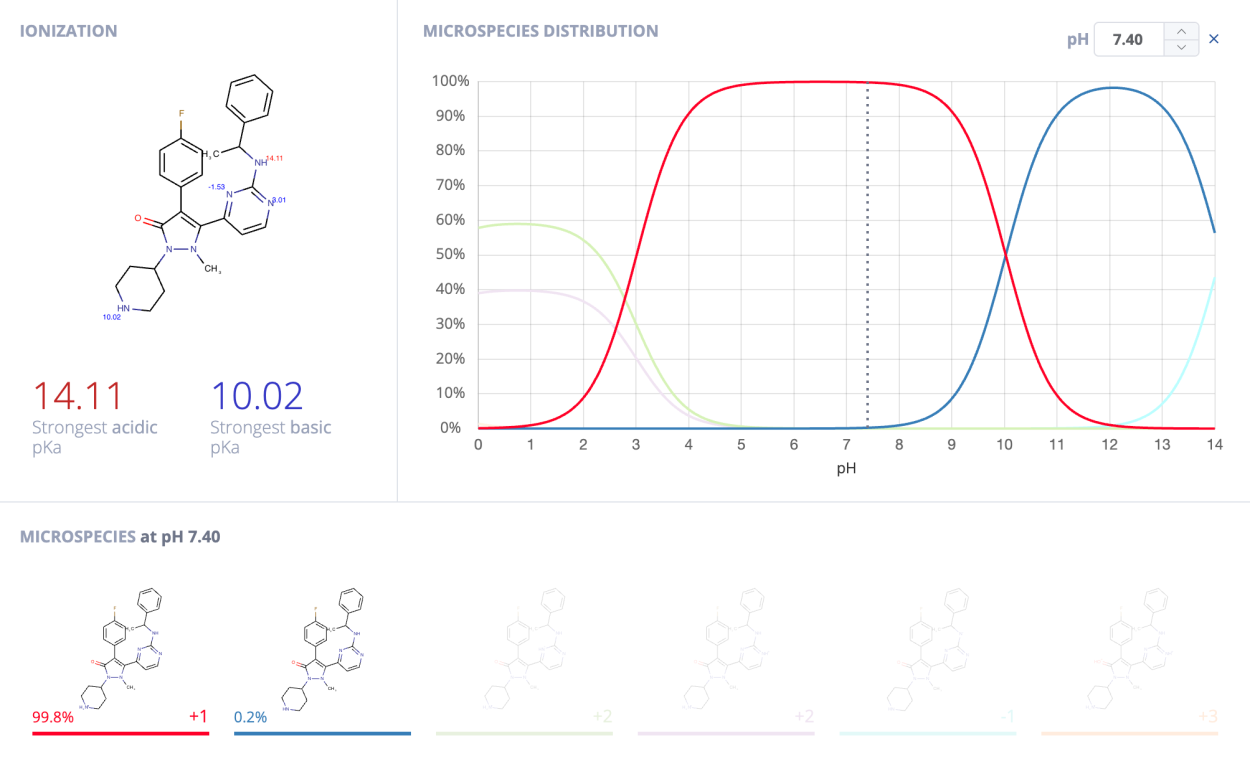
Indeed, the most common microspecies at physiological pH (7.4) according to ChemAxon’s protonation calculator is the one found in the Astex Diverse set.
Rather than manually checking each molecule, we suggest using a command line protonation model, for example the one in the open-source toolkit Open Babel3, to prepare the molecules before loading them with the RDKit. As a last step, we can optimize the hydrogen positions using a force field optimization.
References
Hartshorn, M. J. et al. Diverse, high-quality test set for the validation of protein−ligand docking performance. J. Med. Chem. 50, 726–741 (2007). ↩
RDKit: Open-source cheminformatics. https://www.rdkit.org. ↩
OpenBabel. [https://openbabel.org/] ↩
